Manage a pathway registry#
Background#
Pathways in single-cell analysis represent the interconnected networks of molecular signaling cascades that govern critical cellular processes. They are of utmost importance as they offer a comprehensive understanding of the intricate regulatory mechanisms underlying cellular behavior, providing insights into disease pathogenesis, therapeutic responses, and the identification of potential targets for precision medicine and intervention strategies.
Managing pathways across different datasets is crucial in a biotech company to gain a comprehensive understanding of complex biological processes and facilitate efficient research and development.
In this notebook we are registering the 2023 GO Biological process pathway ontology with Lamin. Afterwards, we are linking the pathways to genes and conducting a pathway enrichment analysis on an interferon-beta treated dataset. Finally, we will demonstrate how to fetch datasets with pathway queries using Lamin.
Setup#
Warning
Please ensure that you have created or loaded a LaminDB instance before running the remaining part of this notebook!
# A lamindb instance containing bionty schema (skip if you already loaded your instance)
!lamin init --storage ./enrichr --schema bionty
Show code cell output
💡 creating schemas: core==0.46.1 bionty==0.30.0
✅ saved: User(id='DzTjkKse', handle='testuser1', email='testuser1@lamin.ai', name='Test User1', updated_at=2023-08-28 13:52:32)
✅ saved: Storage(id='0cFpsy8K', root='/home/runner/work/lamin-usecases/lamin-usecases/docs/enrichr', type='local', updated_at=2023-08-28 13:52:32, created_by_id='DzTjkKse')
✅ loaded instance: testuser1/enrichr
💡 did not register local instance on hub (if you want, call `lamin register`)
import lamindb as ln
import lnschema_bionty as lb
lb.settings.species = "human" # globally set species
import gseapy as gp
import scanpy as sc
import matplotlib.pyplot as plt
from lamin_usecases import datasets as ds
lb.settings.species = "human" # globally set species
✅ loaded instance: testuser1/enrichr (lamindb 0.51.0)
✅ set species: Species(id='uHJU', name='human', taxon_id=9606, scientific_name='homo_sapiens', updated_at=2023-08-28 13:52:34, bionty_source_id='7BBR', created_by_id='DzTjkKse')
✅ set species: Species(id='uHJU', name='human', taxon_id=9606, scientific_name='homo_sapiens', updated_at=2023-08-28 13:52:34, bionty_source_id='7BBR', created_by_id='DzTjkKse')
Fetch GO_Biological_Process_2023 pathways annotated with human genes using Enrichr#
First we fetch the “GO_Biological_Process_2023” pathways for humans using GSEApy which wraps GSEA and Enrichr.
go_bp = gp.get_library(name="GO_Biological_Process_2023", organism="Human")
print(f"Number of pathways {len(go_bp)}")
Number of pathways 5406
go_bp["ATF6-mediated Unfolded Protein Response (GO:0036500)"]
['MBTPS1', 'MBTPS2', 'XBP1', 'ATF6B', 'DDIT3', 'CREBZF']
Parse out the ontology_id from keys, convert into the format of {ontology_id: (name, genes)}
def parse_ontology_id_from_keys(key):
"""Parse out the ontology id.
"ATF6-mediated Unfolded Protein Response (GO:0036500)" -> ("GO:0036500", "ATF6-mediated Unfolded Protein Response")
"""
id = key.split(" ")[-1].replace("(", "").replace(")", "")
name = key.replace(f" ({id})", "")
return (id, name)
go_bp_parsed = {}
for key, genes in go_bp.items():
id, name = parse_ontology_id_from_keys(key)
go_bp_parsed[id] = (name, genes)
go_bp_parsed["GO:0036500"]
('ATF6-mediated Unfolded Protein Response',
['MBTPS1', 'MBTPS2', 'XBP1', 'ATF6B', 'DDIT3', 'CREBZF'])
Register pathway ontology in LaminDB#
pathway_bionty = lb.Pathway.bionty() # equals to bionty.Pathway()
pathway_bionty
Pathway
Species: all
Source: go, 2023-05-10
#terms: 47514
📖 Pathway.df(): ontology reference table
🔎 Pathway.lookup(): autocompletion of terms
🎯 Pathway.search(): free text search of terms
✅ Pathway.validate(): strictly validate values
🧐 Pathway.inspect(): full inspection of values
👽 Pathway.standardize(): convert to standardized names
🪜 Pathway.diff(): difference between two versions
🔗 Pathway.ontology: Pronto.Ontology object
Next, we register all the pathways and genes in LaminDB to finally link pathways to genes.
Register pathway terms#
To register the pathways we make use of .from_values to directly parse the annotated GO pathway ontology IDs into LaminDB.
pathway_records = lb.Pathway.from_values(go_bp_parsed.keys(), lb.Pathway.ontology_id)
✅ created 5406 Pathway records from Bionty matching ontology_id: GO:0044208, GO:0051084, GO:0006103, GO:0061158, GO:0070935, GO:0050427, GO:0042791, GO:0009452, GO:0036261, GO:0006370, GO:0015866, GO:0006167, GO:0046033, GO:0036500, GO:0006754, GO:0046034, GO:0042773, GO:0015867, GO:0086016, GO:0086067, ...
lb.Pathway.from_bionty(ontology_id="GO:0015868")
✅ created 1 Pathway record from Bionty matching ontology_id: GO:0015868
Pathway(id='SMqshx3Y', name='purine ribonucleotide transport', ontology_id='GO:0015868', description='The Directed Movement Of A Purine Ribonucleotide, Any Compound Consisting Of A Purine Ribonucleoside (A Purine Organic Base Attached To A Ribose Sugar) Esterified With (Ortho)Phosphate, Into, Out Of Or Within A Cell.', bionty_source_id='jnW4', created_by_id='DzTjkKse')
ln.save(pathway_records, parents=False) # not recursing through parents
Register gene symbols#
Similarly, we use .from_values for all Pathway associated genes to register them with LaminDB.
all_genes = {g for genes in go_bp.values() for g in genes}
gene_records = lb.Gene.from_values(all_genes, lb.Gene.symbol)
Show code cell output
💡 using global setting species = human
✅ created 14620 Gene records from Bionty matching symbol: KARS1, NAGS, SLC16A9, PAX9, ENTPD4, TCAF1, NDUFAF8, SAR1B, CPSF2, BTF3P11, ZRSR2P1, BTBD2, RCAN2, SCN5A, DENND5A, TMED3, AFG3L2, H2BC8, PDZD7, TIMM8B, ...
✅ created 40 Gene records from Bionty matching synonyms: C1ORF112, C3ORF33, C2ORF49, SLC9A3R1, C1ORF131, TRB, C6ORF15, C1ORF43, C1ORF146, C10ORF90, C6ORF89, C15ORF62, C11ORF80, C17ORF97, C17ORF75, C21ORF91, C12ORF50, C9ORF72, C19ORF12, C12ORF4, ...
❗ ambiguous validation in Bionty for 1082 records: CARS1, DUSP16, SP7, OR2T10, PSMB3, ADAM32, OR2I1P, TAS2R31, DHX36, BDH1, PANK4, GNL1, SALL3, SLC9C1, TRARG1, MDC1, ADGRL1, ZNF707, DUX4L2, NEU4, ...
❗ did not create Gene records for 37 non-validated symbols: AFD1, AZF1, CCL4L1, DGS2, DUX3, DUX5, FOXL3-OT1, IGL, LOC100653049, LOC102723475, LOC102723996, LOC102724159, LOC107984156, LOC112268384, LOC122319436, LOC122513141, LOC122539214, LOC344967, MDRV, MTRNR2L1, ...
gene_records[:3]
[Gene(id='AoCQIhxFvpKD', symbol='KARS1', ensembl_gene_id='ENSG00000065427', ncbi_gene_ids='3735', biotype='protein_coding', description='lysyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:6215]', synonyms='KARS|KARS2|DFNB89', species_id='uHJU', bionty_source_id='AXya', created_by_id='DzTjkKse'),
Gene(id='LyTjAFSrQ9Aj', symbol='NAGS', ensembl_gene_id='ENSG00000161653', ncbi_gene_ids='162417', biotype='protein_coding', description='N-acetylglutamate synthase [Source:HGNC Symbol;Acc:HGNC:17996]', synonyms='NAT7|AGAS|ARGA', species_id='uHJU', bionty_source_id='AXya', created_by_id='DzTjkKse'),
Gene(id='F0sYbSYSynKF', symbol='SLC16A9', ensembl_gene_id='ENSG00000165449', ncbi_gene_ids='220963', biotype='protein_coding', description='solute carrier family 16 member 9 [Source:HGNC Symbol;Acc:HGNC:23520]', synonyms='MCT9|FLJ43803|C10ORF36', species_id='uHJU', bionty_source_id='AXya', created_by_id='DzTjkKse')]
ln.save(gene_records);
Link pathway to genes#
Now that we are tracking all pathways and genes records, we can link both of them to make the pathways even more queryable.
gene_records_ids = {record.symbol: record for record in gene_records}
for pathway_record in pathway_records:
pathway_genes = go_bp_parsed.get(pathway_record.ontology_id)[1]
pathway_genes_records = [gene_records_ids.get(gene) for gene in pathway_genes]
pathway_record.genes.set(pathway_genes_records)
Now genes are linked to pathways:
pathway_record.genes.list("symbol")
['XIAP', 'CAST', 'CARD8', 'CARD18', 'CST7']
A interferon-beta treated dataset#
We will now conduct a pathway enrichment analysis on a small peripheral blood mononuclear cell dataset that is split into control and stimulated groups. The stimulated group was treated with interferon beta.
The dataset was initially obtained using From "SeuratData::ifnb".
Let’s load the dataset and look at the cell type annotations.
adata = ds.anndata_seurat_ifnb()
adata
AnnData object with n_obs × n_vars = 13999 × 14053
obs: 'orig.ident', 'nCount_RNA', 'nFeature_RNA', 'stim', 'seurat_annotations'
var: 'features'
uns: 'log1p'
adata.obs["seurat_annotations"].value_counts()
seurat_annotations
CD14 Mono 4362
CD4 Naive T 2504
CD4 Memory T 1762
CD16 Mono 1044
B 978
CD8 T 814
NK 633
T activated 619
DC 472
B Activated 388
Mk 236
pDC 132
Eryth 55
Name: count, dtype: int64
For simplicity, we subset to “B Activated” cells:
adata_ba = adata[adata.obs.seurat_annotations == "B Activated"].copy()
adata_ba
AnnData object with n_obs × n_vars = 388 × 14053
obs: 'orig.ident', 'nCount_RNA', 'nFeature_RNA', 'stim', 'seurat_annotations'
var: 'features'
uns: 'log1p'
Pathway enrichment analysis using Enrichr#
This analysis is based on: https://gseapy.readthedocs.io/en/master/singlecell_usecase.html
First, we compute differentially expressed genes using a Wilcoxon test between stimulated and control cells.
# compute differentially expressed genes
sc.tl.rank_genes_groups(
adata_ba,
groupby="stim",
use_raw=False,
method="wilcoxon",
groups=["STIM"],
reference="CTRL",
)
rank_genes_groups_df = sc.get.rank_genes_groups_df(adata_ba, "STIM")
rank_genes_groups_df.head()
| names | scores | logfoldchanges | pvals | pvals_adj | |
|---|---|---|---|---|---|
| 0 | ISG15 | 16.881584 | 5.923428 | 6.147295e-64 | 6.536230e-60 |
| 1 | ISG20 | 16.857113 | 4.167713 | 9.302256e-64 | 6.536230e-60 |
| 2 | IFIT3 | 14.587233 | 31.232290 | 3.386569e-48 | 1.586382e-44 |
| 3 | IFI6 | 14.128634 | 6.471180 | 2.530019e-45 | 8.888589e-42 |
| 4 | MX1 | 13.442097 | 6.241539 | 3.425901e-41 | 9.628837e-38 |
Next, we filter out up/down-regulated differentially expressed gene sets:
degs_up = rank_genes_groups_df[
(rank_genes_groups_df["logfoldchanges"] > 0)
& (rank_genes_groups_df["pvals_adj"] < 0.05)
]
degs_dw = rank_genes_groups_df[
(rank_genes_groups_df["logfoldchanges"] < 0)
& (rank_genes_groups_df["pvals_adj"] < 0.05)
]
degs_up.shape, degs_dw.shape
((89, 5), (47, 5))
Run pathway enrichment analysis on DEGs and plot top 10 pathways:
enr_up = gp.enrichr(degs_up.names, gene_sets="GO_Biological_Process_2023").res2d
gp.dotplot(enr_up, figsize=(2, 3), title="Up", cmap=plt.cm.autumn_r);

enr_dw = gp.enrichr(degs_dw.names, gene_sets="GO_Biological_Process_2023").res2d
gp.dotplot(enr_dw, figsize=(2, 3), title="Down", cmap=plt.cm.winter_r, size=10);
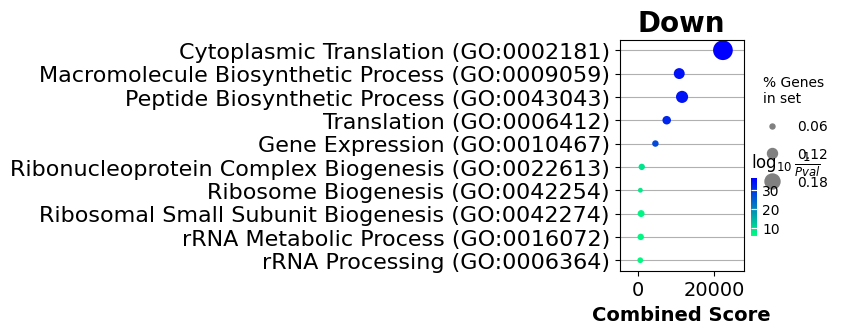
Track datasets containing annotated pathways in LaminDB#
Let’s enable tracking of the current notebook as the transform of this file:
ln.track()
💡 notebook imports: gseapy==1.0.5 lamin_usecases==0.0.1 lamindb==0.51.0 lnschema_bionty==0.30.0 matplotlib==3.7.2 scanpy==1.9.4
✅ saved: Transform(id='6oxEIEduvo6wz8', name='Manage a pathway registry', short_name='enrichr', version='0', type=notebook, updated_at=2023-08-28 13:53:35, created_by_id='DzTjkKse')
✅ saved: Run(id='3Hv5nR06PhFH1sJTMCo5', run_at=2023-08-28 13:53:35, transform_id='6oxEIEduvo6wz8', created_by_id='DzTjkKse')
We further create a File object to track the dataset.
file = ln.File.from_anndata(
adata_ba, description="seurat_ifnb_activated_Bcells", var_ref=lb.Gene.symbol
)
💡 file will be copied to default storage upon `save()` with key `None` ('.lamindb/OtpoPa7zEqdiX9v74f3X.h5ad')
💡 parsing feature names of X stored in slot 'var'
💡 using global setting species = human
✅ 9324 terms (66.30%) are validated for symbol
❗ 4729 terms (33.70%) are not validated for symbol: AL627309.1, RP11-206L10.2, LINC00115, KLHL17, C1orf159, ACAP3, CPSF3L, GLTPD1, RP4-758J18.2, AL645728.1, RP11-345P4.9, SLC35E2B, SLC35E2, RP5-892K4.1, C1orf86, AL590822.2, MORN1, RP3-395M20.12, RP3-395M20.9, FAM213B, ...
💡 using global setting species = human
✅ linked: FeatureSet(id='EfN2XftQOtWX7eoSuYP6', n=10599, type='float', registry='bionty.Gene', hash='ehMHlXCXiKumXLoHU96e', created_by_id='DzTjkKse')
💡 parsing feature names of slot 'obs'
❗ 5 terms (100.00%) are not validated for name: orig.ident, nCount_RNA, nFeature_RNA, stim, seurat_annotations
❗ no validated features, skip creating feature set
ln.save(file)
✅ saved 1 feature set for slot: 'var'
✅ storing file 'OtpoPa7zEqdiX9v74f3X' at '.lamindb/OtpoPa7zEqdiX9v74f3X.h5ad'
We further create two feature sets for degs_up and degs_dw which we can later associate with the associated pathways:
degs_up_featureset = ln.FeatureSet.from_values(degs_up.names, lb.Gene.symbol)
Show code cell output
💡 using global setting species = human
✅ 76 terms (85.40%) are validated for symbol
❗ 13 terms (14.60%) are not validated for symbol: EPSTI1, WARS, LAP3, SAMD9L, NMI, TMEM123, CMPK2, H3F3B, PSMA2.1, PHF11, CLEC2D, DDX58, CD48
💡 using global setting species = human
degs_dw_featureset = ln.FeatureSet.from_values(degs_dw.names, lb.Gene.symbol)
Show code cell output
💡 using global setting species = human
✅ 44 terms (93.60%) are validated for symbol
❗ 3 terms (6.40%) are not validated for symbol: GNB2L1, TMEM66, HLA-DQB1
💡 using global setting species = human
Link the top 10 pathways to the corresponding differentially expressed genes:
# get ontology ids for the top 10 pathways
enr_up_top10 = [
pw_id[0] for pw_id in enr_up.head(10).Term.apply(parse_ontology_id_from_keys)
]
enr_dw_top10 = [
pw_id[0] for pw_id in enr_dw.head(10).Term.apply(parse_ontology_id_from_keys)
]
# get pathway records
enr_up_top10_pathways = lb.Pathway.from_values(enr_up_top10, lb.Pathway.ontology_id)
enr_dw_top10_pathways = lb.Pathway.from_values(enr_dw_top10, lb.Pathway.ontology_id)
Link feature sets to file:
file.features.add_feature_set(degs_up_featureset, slot="up-DEGs")
file.features.add_feature_set(degs_dw_featureset, slot="down-DEGs")
Associate the pathways to the differentially expressed genes:
degs_up_featureset.pathways.set(enr_up_top10_pathways)
degs_dw_featureset.pathways.set(enr_dw_top10_pathways)
degs_up_featureset.pathways.list("name")
['positive regulation of interferon-beta production',
'response to type II interferon',
'response to interferon-beta',
'defense response to virus',
'defense response to symbiont',
'negative regulation of viral process',
'negative regulation of viral genome replication',
'interleukin-27-mediated signaling pathway',
'antiviral innate immune response',
'regulation of viral genome replication']
Querying for pathways#
Querying for pathways is now simple with .filter:
lb.Pathway.filter(name__contains="interferon-beta").df()
| name | ontology_id | abbr | synonyms | description | bionty_source_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| SGYMKD7O | positive regulation of interferon-beta production | GO:0032728 | None | positive regulation of IFN-beta production|up-... | Any Process That Activates Or Increases The Fr... | jnW4 | 2023-08-28 13:52:53 | DzTjkKse |
| uu9GYFx2 | negative regulation of interferon-beta production | GO:0032688 | None | down regulation of interferon-beta production|... | Any Process That Stops, Prevents, Or Reduces T... | jnW4 | 2023-08-28 13:52:53 | DzTjkKse |
| mCgM7JYR | response to interferon-beta | GO:0035456 | None | response to fiblaferon|response to fibroblast ... | Any Process That Results In A Change In State ... | jnW4 | 2023-08-28 13:52:53 | DzTjkKse |
| l06ZujxW | cellular response to interferon-beta | GO:0035458 | None | cellular response to fibroblast interferon|cel... | Any Process That Results In A Change In State ... | jnW4 | 2023-08-28 13:52:53 | DzTjkKse |
| GD9xCHBK | regulation of interferon-beta production | GO:0032648 | None | regulation of IFN-beta production | Any Process That Modulates The Frequency, Rate... | jnW4 | 2023-08-28 13:52:53 | DzTjkKse |
Query pathways from a gene:
lb.Pathway.filter(genes__symbol="KIR2DL1").df()
| name | ontology_id | abbr | synonyms | description | bionty_source_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|
| id | ||||||||
| TSXmNUbN | immune response-inhibiting cell surface recept... | GO:0002767 | None | immune response-inhibiting cell surface recept... | The Series Of Molecular Signals Initiated By A... | jnW4 | 2023-08-28 13:52:54 | DzTjkKse |
Query files from a pathway:
ln.File.filter(feature_sets__pathways__name__icontains="interferon-beta").first()
File(id='OtpoPa7zEqdiX9v74f3X', suffix='.h5ad', accessor='AnnData', description='seurat_ifnb_activated_Bcells', size=5896640, hash='vAWd5emmLj0nv0E0x5LOSA', hash_type='md5', updated_at=2023-08-28 13:53:37, storage_id='0cFpsy8K', transform_id='6oxEIEduvo6wz8', run_id='3Hv5nR06PhFH1sJTMCo5', created_by_id='DzTjkKse')
Query featuresets from a pathway to learn from which geneset this pathway was computed:
pathway = lb.Pathway.filter(ontology_id="GO:0035456").one()
pathway
Pathway(id='mCgM7JYR', name='response to interferon-beta', ontology_id='GO:0035456', synonyms='response to fiblaferon|response to fibroblast interferon|response to interferon beta', description='Any Process That Results In A Change In State Or Activity Of A Cell Or An Organism (In Terms Of Movement, Secretion, Enzyme Production, Gene Expression, Etc.) As A Result Of An Interferon-Beta Stimulus. Interferon-Beta Is A Type I Interferon.', updated_at=2023-08-28 13:52:53, bionty_source_id='jnW4', created_by_id='DzTjkKse')
degs = ln.FeatureSet.filter(pathways__ontology_id=pathway.ontology_id).one()
Now we can get the list of genes that are differentially expressed and belong to this pathway:
pathway_genes = set(pathway.genes.list("symbol"))
degs_genes = set(degs.genes.list("symbol"))
pathway_genes.intersection(degs_genes)
{'BST2',
'IFI16',
'IFITM2',
'IFITM3',
'IRF1',
'OAS1',
'PLSCR1',
'STAT1',
'XAF1'}
Conclusion#
Registering pathways and associated gene sets is made simple with .from_values that ensures that all parsed objects are linked to ontology IDs.Linking both sets is possible with FeatureSet to facilitate simple querying for datasets that contain specific pathways. Since the pathways are linked to genes, Lamin also enables fetching the associated genes of a registered pathway to, for usecase, retrieve sets of differentially expressed genes that are a part of a specific pathway.
Try it yourself#
This notebook is available at laminlabs/lamin-usecases.
Show code cell content
!lamin delete --force enrichr
!rm -r ./enrichr
💡 deleting instance testuser1/enrichr
✅ deleted instance settings file: /home/runner/.lamin/instance--testuser1--enrichr.env
✅ instance cache deleted
✅ deleted '.lndb' sqlite file
❗ consider manually deleting your stored data: /home/runner/work/lamin-usecases/lamin-usecases/docs/enrichr
